 Time-space registration is
what you do all your waking time. Objects move, shake, jump,
but it does not prevent you from recognizing & understanding their characteristics.
The brain does the registration for you. For example, let's mimic what your eyes might see
when you observe living cells through a microscope. (Movies below show epithelial cells with karotin).
On the top left panel you see image distructed by a motion: the cells are moving,
the microscope or the table is shaking.
Can we make the movie steady without altering the image quality?
Yes, we can. I wrote a Matlab registration algorithm that determines how far to shift each image
in order to make the whole movie steady. The top right panel shows the
result of the registration algorithm: the "calm" cells.
There are 10 frames in the movie, as 10 consequtive microscope images.
Time-space registration is
what you do all your waking time. Objects move, shake, jump,
but it does not prevent you from recognizing & understanding their characteristics.
The brain does the registration for you. For example, let's mimic what your eyes might see
when you observe living cells through a microscope. (Movies below show epithelial cells with karotin).
On the top left panel you see image distructed by a motion: the cells are moving,
the microscope or the table is shaking.
Can we make the movie steady without altering the image quality?
Yes, we can. I wrote a Matlab registration algorithm that determines how far to shift each image
in order to make the whole movie steady. The top right panel shows the
result of the registration algorithm: the "calm" cells.
There are 10 frames in the movie, as 10 consequtive microscope images.
acquired data
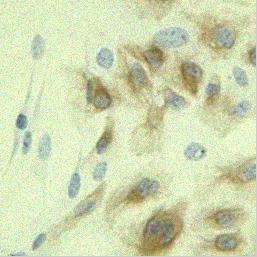 |
registered data
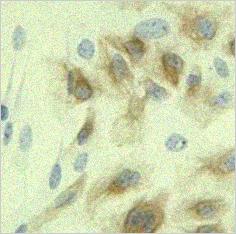 |
|
Now, lets imagine a more realistic scenario, at each acquisition the microscope
zooms in (or the cell/tissue swells). Thus, in addition to the image shift, there is a non-uniform
image transformation that occured over 15 steps (the lower left panel).
The registration algorithm still works! (the lower right panel).
acquired data
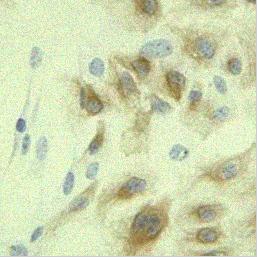 |
registered data
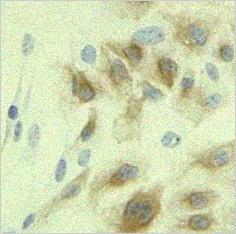 |
|
Each color image consists of 256-by-256-by-3 pixels, which are acquired in 10 time steps,
each time for 15 depths. Therefore, the algorithm works with a 5-D matrix.
 Time-space registration
Time-space registration


